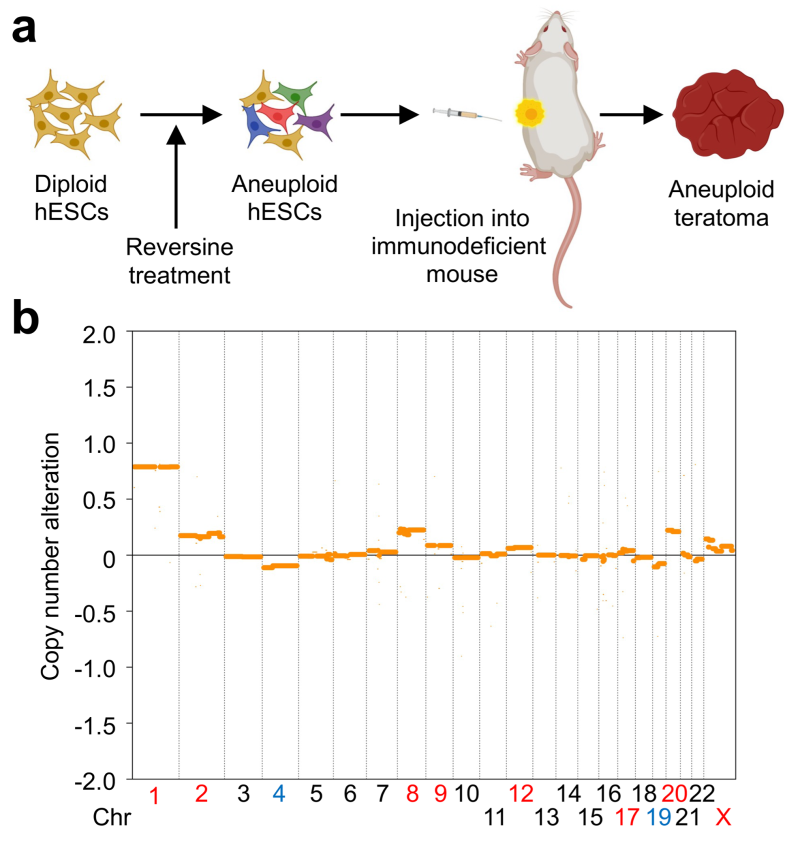
Aneuploidy is a common trait of tumor cells. It is not fully understood how aneuploidy leads to human tumorigenesis. Understanding the evolution and consequences of aneuploidy may reveal common vulnerabilities of aneuploid tumors that can be exploited to treat cancer. By injecting reversine-treated human embryonic stem cells (hESCs) into immunodeficient mice and growing into teratoma, we have successfully created aneuploid tumors from genetically stable diploid cells. In the future, we will study aneuploid teratomas with single-cell multiomics analysis and identify expression profiles and spatial locations of aneuploid cells within the tumor. We will define the molecular characteristics of these engineered aneuploid tumors that enable their fast proliferation despite the proteotoxic stress. Aneuploidy has been implicated in the immune evasion of tumors. We will grow diploid and aneuploid teratomas in humanized mice with a partially reconstituted human immune system. By comparing aneuploid tumors grown in immunodeficient and humanized mice, we will reveal whether and how the immune system exerts selective pressure on the pattern of aneuploidy. Finally, we will perform CRISPR screens with druggable genome sub-libraries or other custom sub-libraries to identify vulnerabilities unique to aneuploid teratomas in immunodeficient or humanized mice. This screen may identify novel molecular targets that, when inhibited, directly kill aneuploid tumor cells or sensitize them to killing by immune cells.
RESEARCH
1
Evolution and consequences of aneuploidy during tumorigenesis
2
Establishment of sister-chromatid cohesion during DNA replication
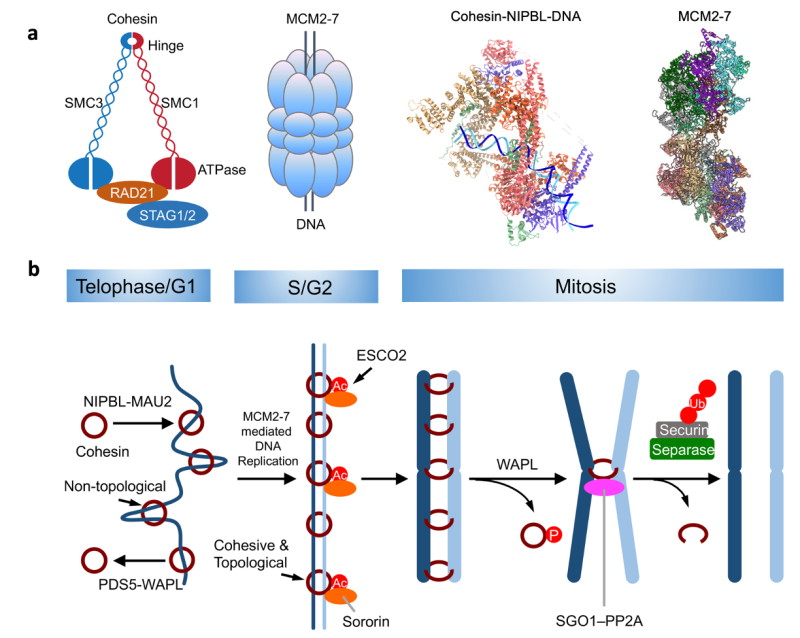
The cohesin ring topologically entraps both sister chromatids, thereby establishing sister-chromatid cohesion. The mechanism of cohesion establishment during DNA replication is not understood. The current evidence suggests that cohesin and NIPBL interact with both DDK-modified MCM2–7 and replisome components bound to the active CMG replicative helicase. The former interaction is specific to metazoans whereas the latter is conserved from yeast to human. We will in vitro reconstitute both interactions with human cohesin, NIPBL, PDS5B, ESCO2 (the cohesin acetyltransferase), sororin, MCM2–7, DDK, and replisome components, and dissect these interactions using standard pulldown and crosslinking/mass spectrometry methods. We will determine the structures of the MCM–ESCO2, MCM–cohesin–NIPBL, replisome–cohesin–NIPBL, and endogenous cohesive cohesin–DNA complexes with cryo-EM. Establishing molecular details of the cohesin–replisome interactions will enable the design of separation-of-function replisome mutants that support DNA replication but not cohesion. We will examine the phenotypes of key mutants in human cells using a depletion-complementation strategy and established replication and cohesion assays. These biochemical, structural, and functional analyses will reveal how cohesive cohesin holds two sister chromatids and how cohesion establishment is coupled to DNA replication.

3
3D genome organization and cell fate decisions
There are about 200 cell types in the human body. All have the same genome. It is the selective expression of specific subsets of genes in pluripotent stem cells that determines the cell fate. We hypothesize that dynamic 3D genome organization makes and breaks long-range interactions between distal DNA elements, including promoters and enhancers. As such, it shapes the gene expression landscape of the genome in response to developmental cues, thereby impacting cell fate decisions. Consistent with this hypothesis, we have recently shown that deletion of the cohesin subunit gene STAG2 in the nervous system causes delayed differentiation of the oligodendrocyte lineage, hypomyelination, and neurological defects. In the future, we will further test this hypothesis using brain organoids derived from hESCs as the model system. By using protein degradation system such as HaloPROTAC or dTAG to deplete CTCF or cohesin subunits from hESCs and developing organoids within hours, we will test whether the depletion of CTCF or cohesin subunits alters the differentiation of certain cell types in the organoids. We will also delete functional DNA elements, such as CTCF sites, to perturb chromosome loops near cell-fate-determining genes in hESCs and establish a requirement of certain loops in cell fate decisions during organoid formation. Further mechanism studies will determine how 3D genome organization regulates gene expression and cell fate decisions.